Our lab is interested in the physical principles underlying vertebrate regeneration. Regeneration is the astonishing process in which an organism regrows a lost body part. We ask how cells communicate and coordinate with each other for a vertebrate body part to regenerate into its proper form. How are signals organized in time and space and integrated with mechanical forces to orchestrate growth and patterning?
We ask these questions with a quantitative approach at the boundary of physics and biology. We apply experimental, computational, and theoretical methods. We monitor regeneration live, at subcellular resolution and tissue-wide, including in its native context. We use high-sensitivity sensors and reporters to visualize signals, forces and cell dynamics and we analyze data using automated quantification methods. We use theory to conceptualize those complex systems, generate hypotheses and guide the interpretation of experimental data.
Our main model system is zebrafish and we are fond of zebrafish scales. Scales are bone disks that, after loss, regenerate in about two weeks. Regeneration is driven by a monolayer of osteoblasts that regrows and deposits the new bone. We like scales because they are accessible to live imaging, simple enough to be studied quantitatively and have an intriguing regenerative biology. Thus, scales are an ideal system to study how mechano-chemical signals coordinate morphogenesis in regeneration.
We have open positions for Master students, PhD students and postdocs who want to work at the boundary of physics and biology to tackle the mechanisms of regeneration. We are looking for biologists, physicists, engineers, mathematicians, computer scientists and more, interested in developing or strengthening an interdisciplinary profile.
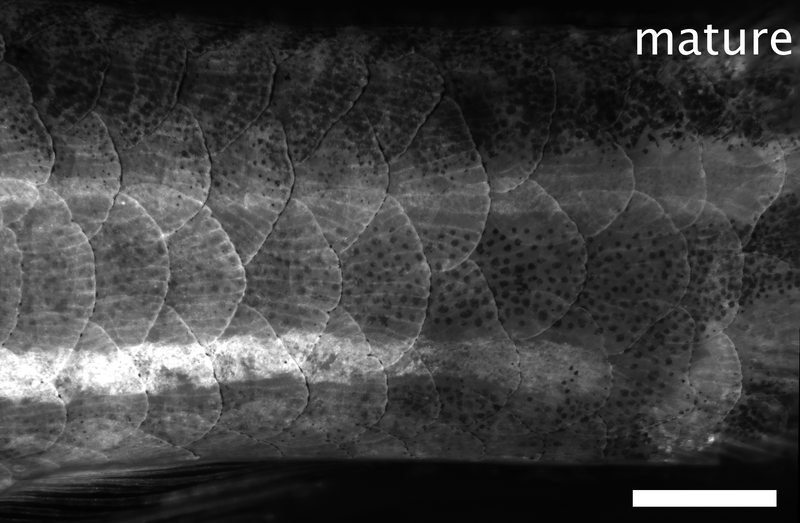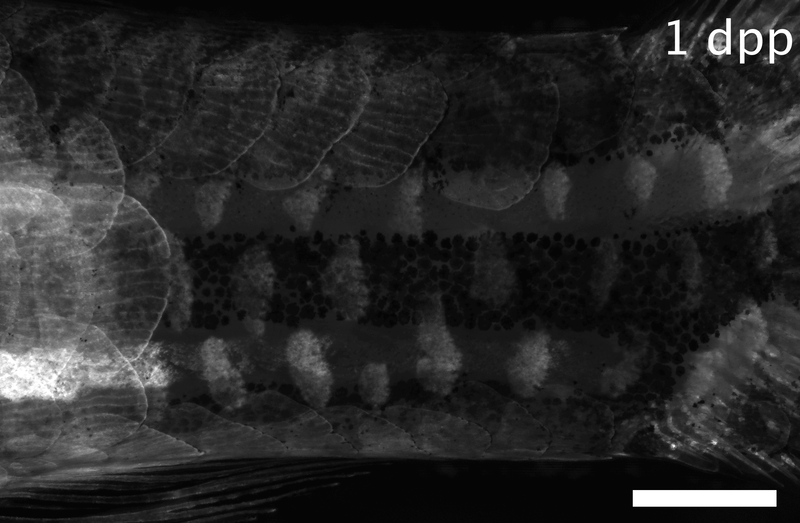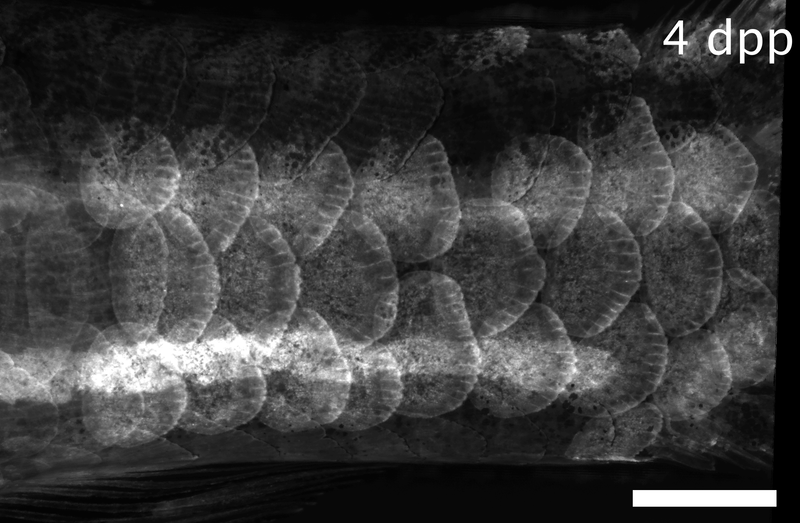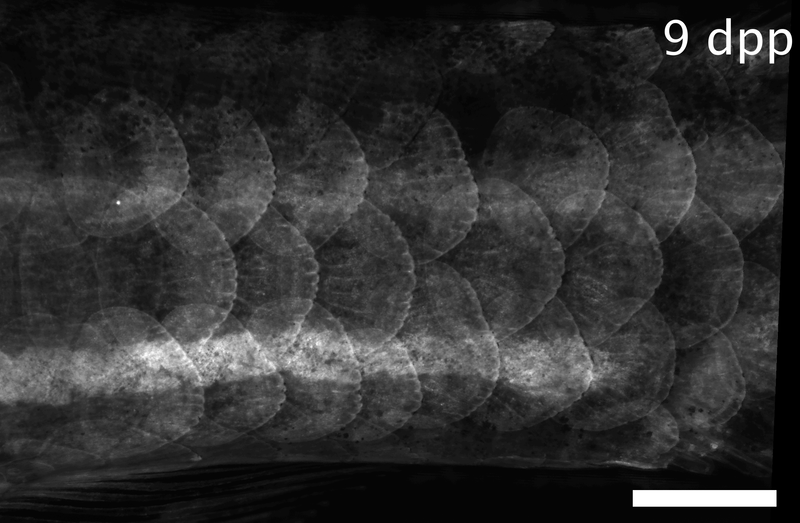
Array of regenerating scales on the trunk of a zebrafish, visualized by the osteoblast marker osx:GFP-CAAX. Dpp: days post-plucking. Scale bar: 1 mm.*
Research projects
Control of scale size and shape by Erk activity waves
How do signals control the final size of a regenerated tissue? In scale regeneration, cell growth is coordinated by Erk activity waves that originate at the center of the scale and travel across the entire tissue. The more waves are generated, the more the scale will grow. In this project, we ask how waves are generated and propagated, so that scales reach their appropriate final size. What mechanism controls wave generation? Is there a feedback mechanism that informs the waves “source” about tissue size, so that wave generation is tuned accordingly?
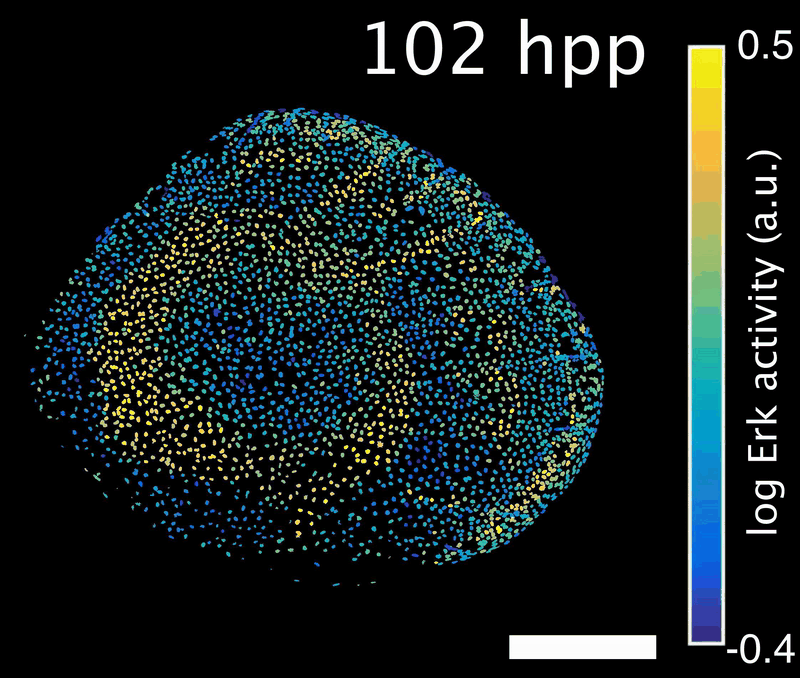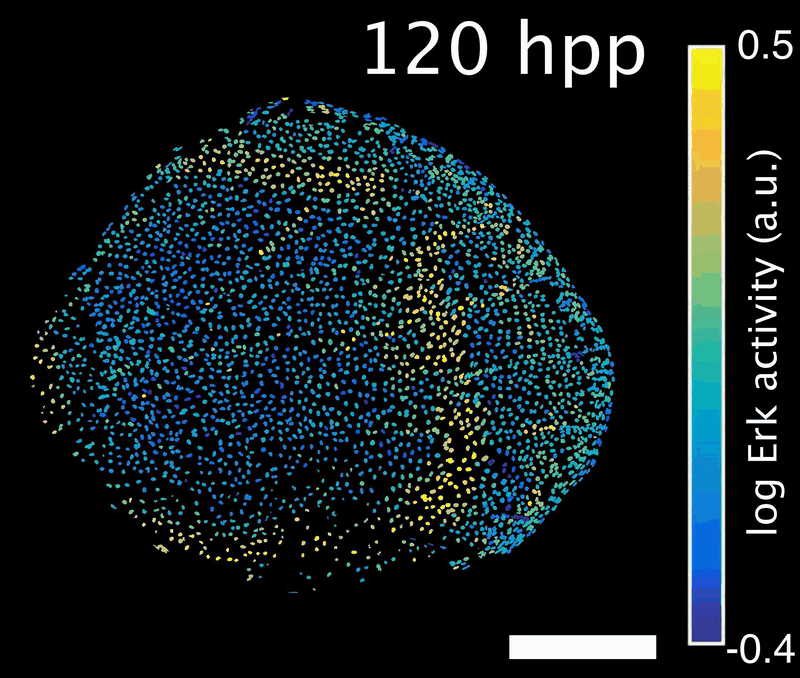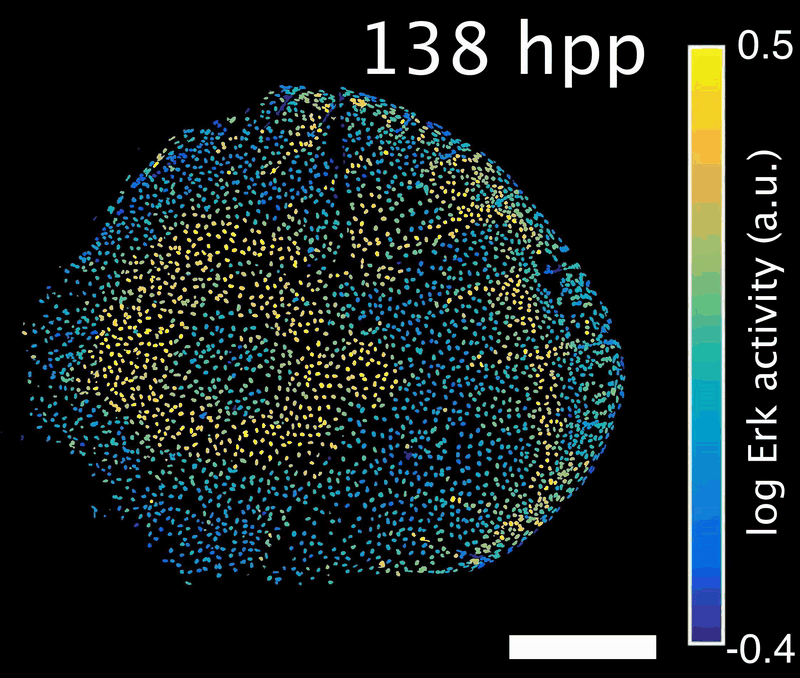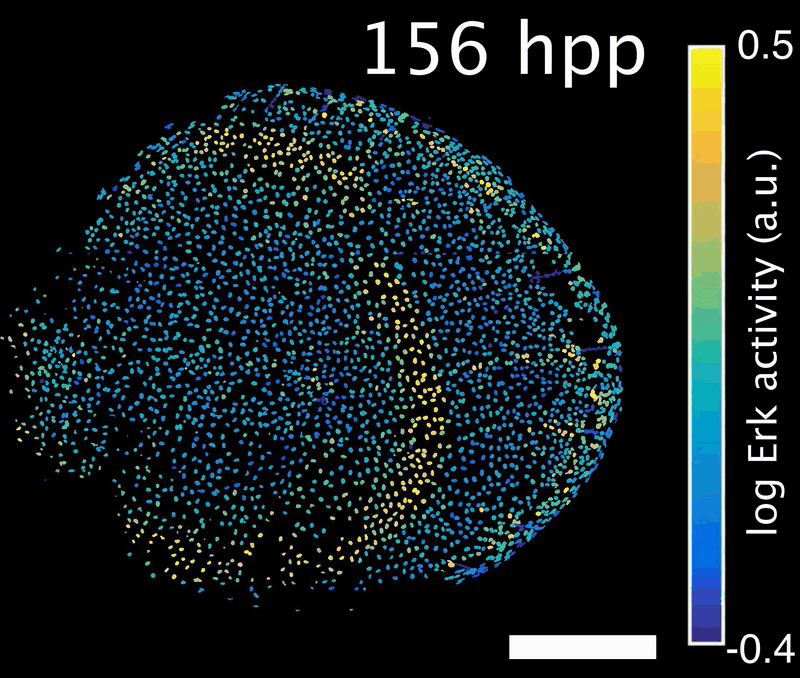
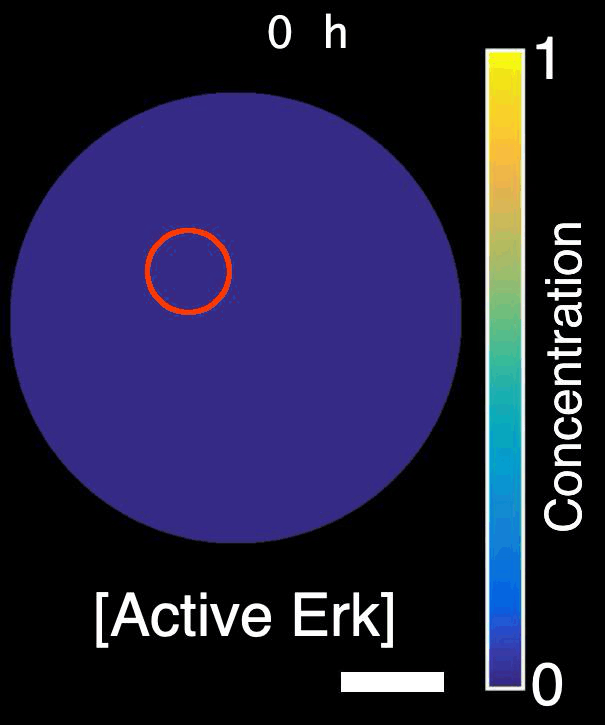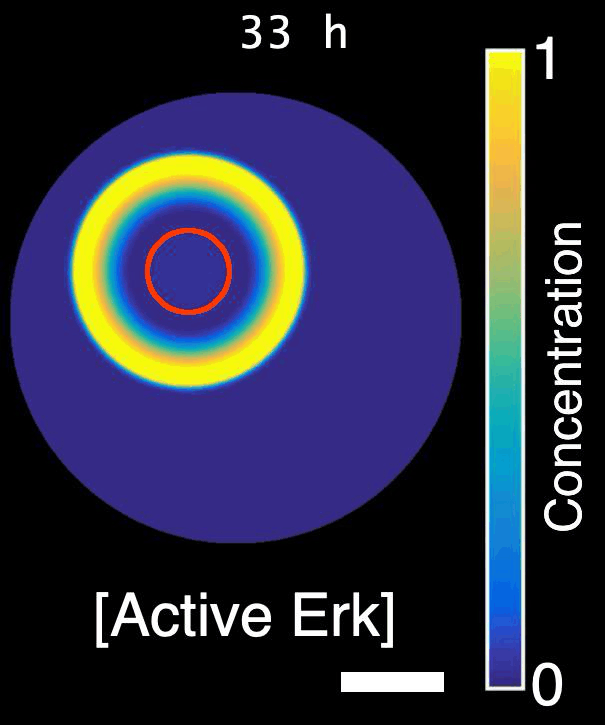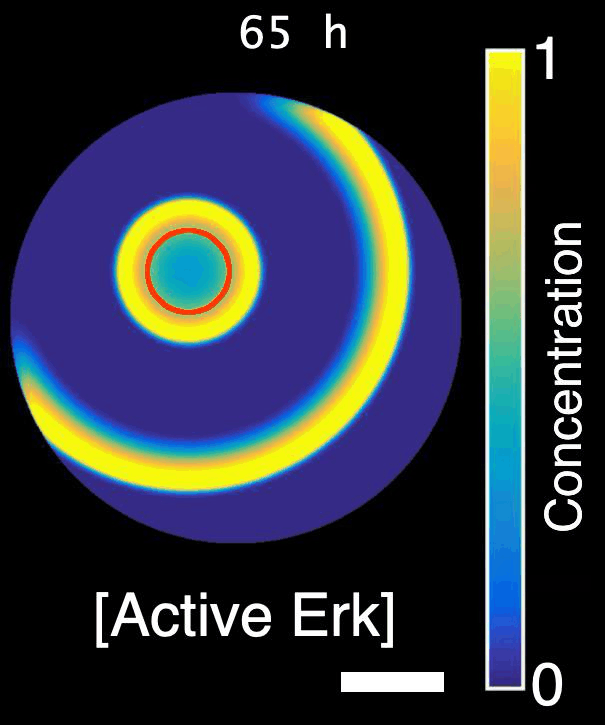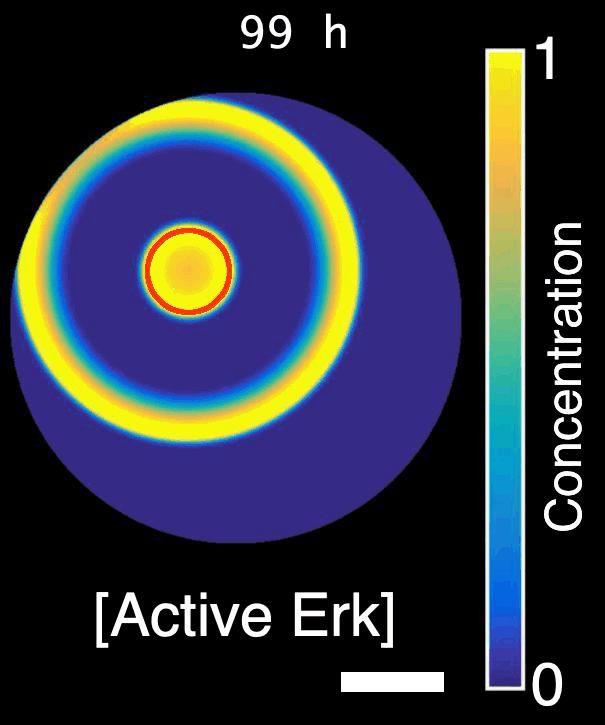
Erk activity waves in a regenerating scale. Hpp: hours post-plucking. Scale bar: 250 µm.*
How are signals propagated in a large regenerating tissue? Erk activity waves are compatible with a reaction-diffusion system involving a diffusible Erk activator such as a growth factor, Erk itself and an Erk inhibitor. In this project, we will ask what the molecular components of this mechanism are and how do they interact with each other to propagate waves.
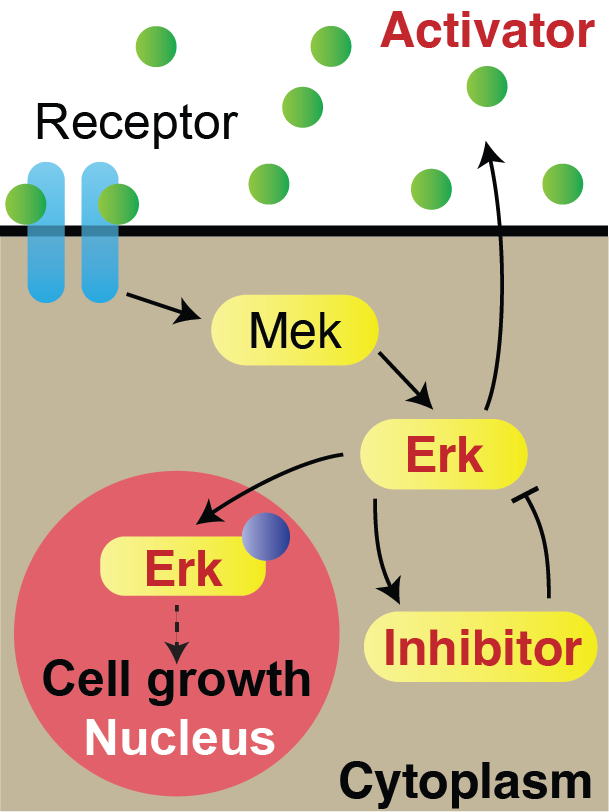
Model of Erk signalling dynamics including a diffusible Erk activator, positive feedback between Erk and the activator, and a negative feedback including an Erk inhibitor. Left: schematics. Right: simulation. Red shaded region: constant source of activator. Scale-bar: 250 µm. *
How does tissue growth lead to form? Erk activity waves impart patterns of tissue growth. Are Erk activity waves a way to distribute growth-induced stresses across the tissues? What are the effects on wave-patterned growth on the final shape of a scale?
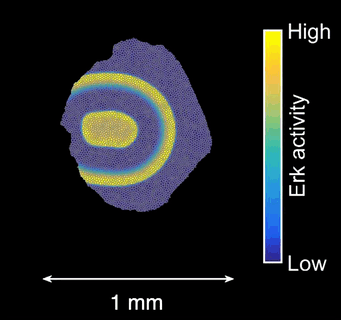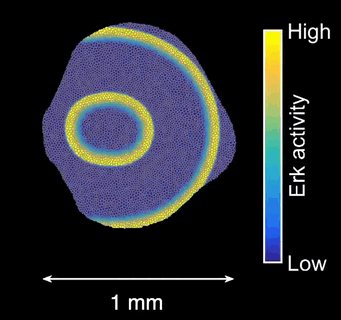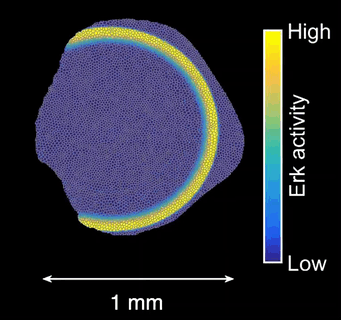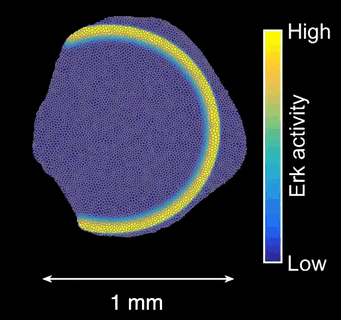
Schematics of scale morphogenesis driven by a train of Erk activity waves.*
Coordination of cell growth and bone formation
How are different cell processes coordinated to obtain a regenerated body part that is properly structured? In scale regeneration, osteoblasts deposit bone matrix while they grow, instructed by Erk activity waves. In this project, we will put together the spatial-temporal detail of live imaging with the comprehensive view of transcriptomics to understand how dynamic signals orchestrate different pathways and cell behaviours in regeneration.
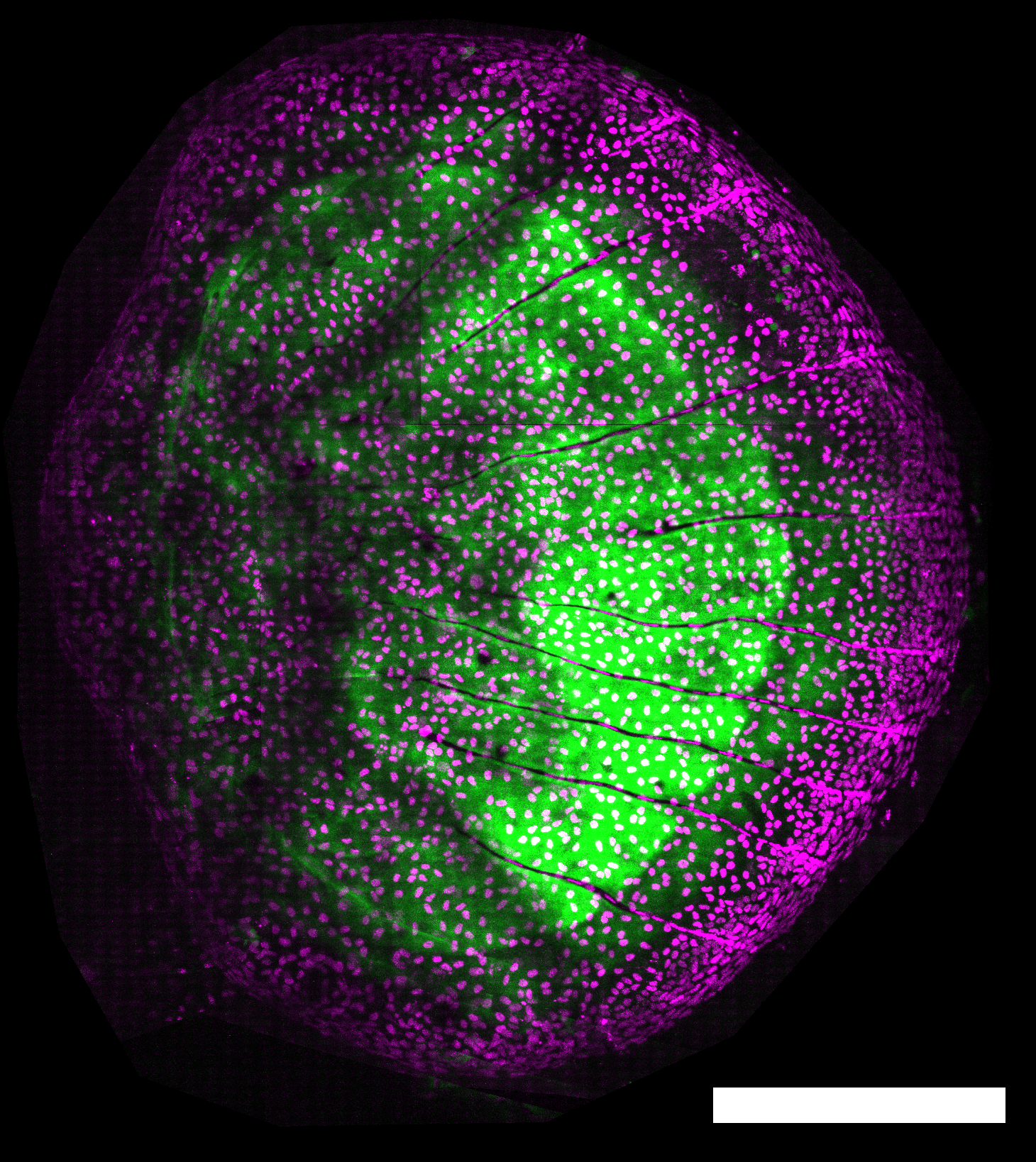
Bone (green) in a regenerating scale (5 days post-plucking; green: Calcein stain; magenta: osx:H2A-mCherry). Scale-bar: 250 µm.
Mechano-chemical coupling in the control of cell proliferation
How is the transition between different phases of regeneration controlled? Put otherwise, how do cells know it is time to change behaviour? During an early phase of scale regeneration, Erk is uniformly active and osteoblasts actively proliferate. Then, Erk switches off in an activation wave that starts at the center of the scale and propagates out. At the same time, osteoblast transition from proliferation to growth without cell division (hypertrophy). In this project, we ask how Erk is patterned over time to control the transition from proliferation to hypertrophy. Our overarching hypothesis is that Erk integrates mechanical inputs from within the tissue and from the neighboring bone. We are relating Erk dynamics, forces, bone biophysical properties and osteoblast behaviour to understand how their interplay controls the final number of cells in a regenerating scale.
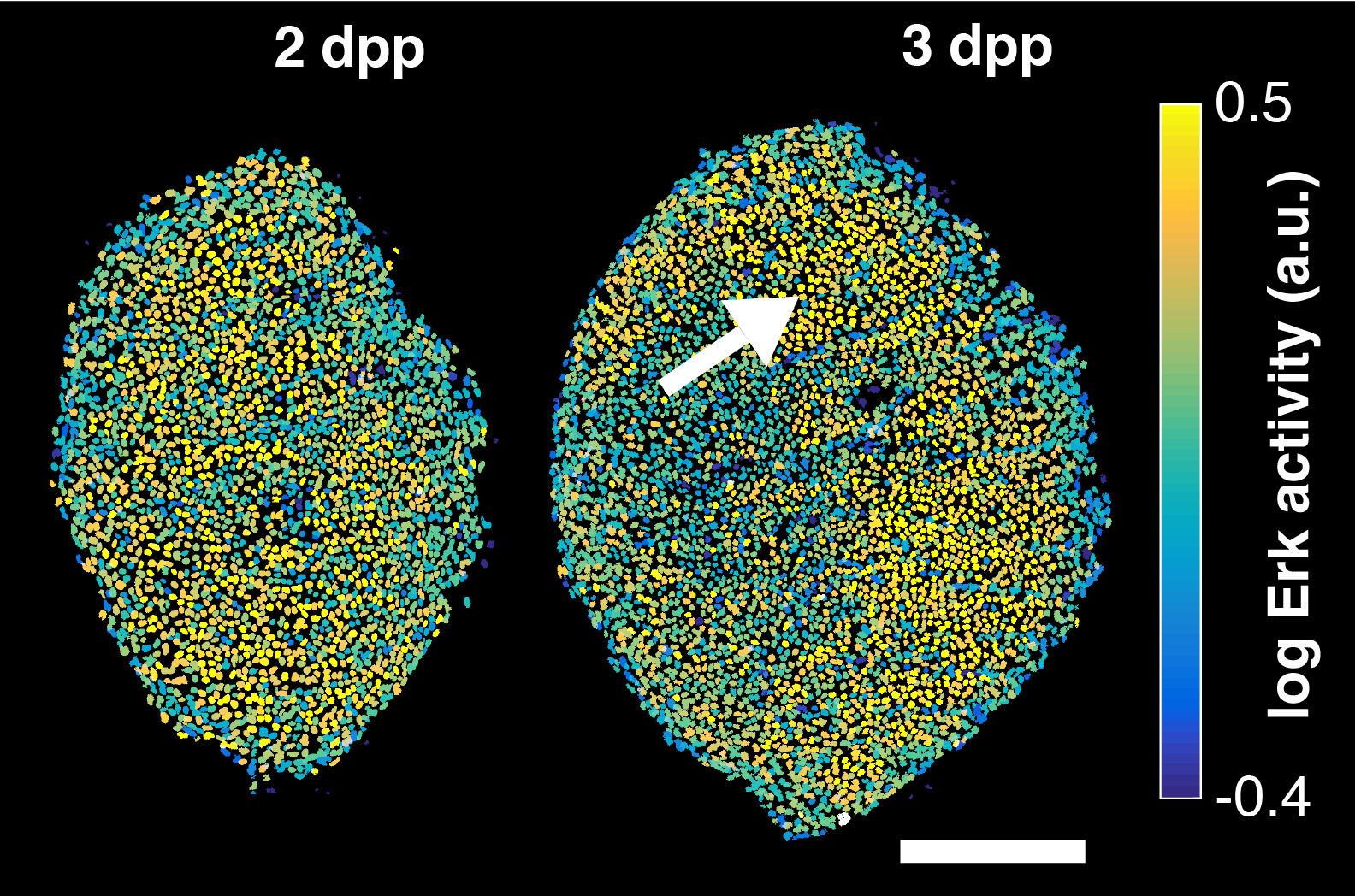
Example of Erk activity in a regenerating scale at 2 and 3 days post-plucking (dpp). Erk activity is activated in a uniform pattern and then switches off starting from the scale centre. Scale-bar: 250 µm. *
*: Figure adapted from De Simone et al., Nature, 2021, DOI: 10.1038/s41586-020-03085-8; Copyright © 2021, The Author(s), under exclusive license to Springer Nature Limited.
External funding
- SNSF Eccellenza Professorial Fellowship PCEFP3_202776
- ERC Starting Grant 2021 (funded by SERI)
