- news
- 26-04-2021
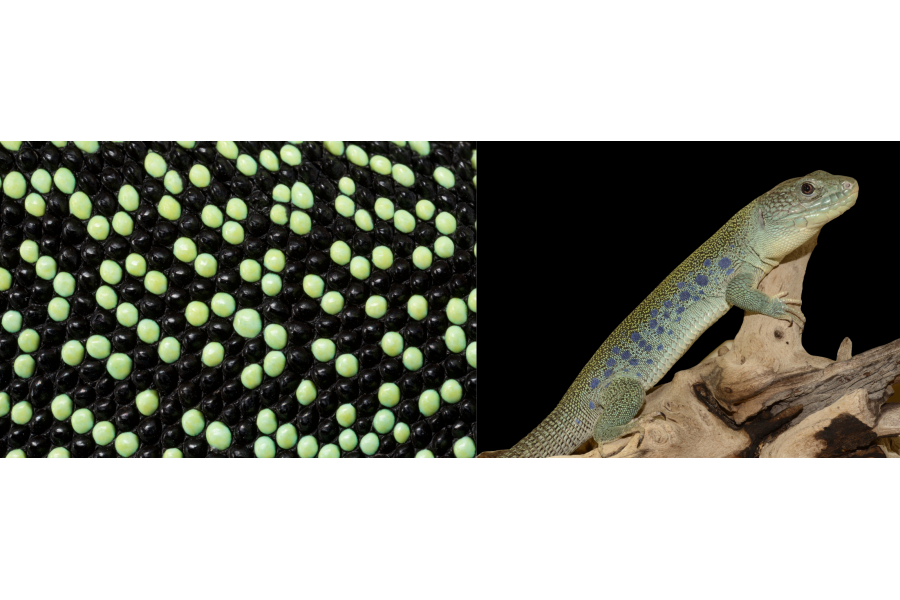
The Milinkovitch's lab uses RD numerical simulations in 3D on realistic lizard skin geometries and demonstrates that skin thickness variation on its own is sufficient to cause scale-by-scale coloration and CA dynamics during RD patterning. In addition, they show that this phenomenon is robust to RD model variation. Finally, the team shows that animal growth affects the scale-colour flipping dynamics.
The Milinkovitch team previously showed in Nature (2017: vol. 544, 173–179) that the adult ocellated lizard skin colour pattern is effectively generated by a stochastic cellular automaton (CA) of skin scales. They additionally suggested that the canonical continuous two-dimensional (2D) reaction-diffusion (RD) process of colour pattern development is transformed into this discrete CA because of the reduced diffusion coefficients at the borders of scales (justified by the corresponding thinning of the skin). However, as all their previous RD simulations included only two spatial dimensions (2D), the entirety of their demonstration relied on the explicit assumption that variations in the third spatial dimension of the ocellated lizard’s skin (i.e., the presence of thick skin scales with much thinner borders) justifies a substantial reduction of the continuous RD diffusion coefficients at the one-dimensional edges of 2D scales, effectively decoupling the RD dynamics inside versus between scales.
In their new publication (Nature Communications 2021), the team now tests the validity of this key assumption by performing RD numerical simulations in a three-dimensional (3D) domain of skin scales such that they ‘let the 3D geometry do the job’, i.e., they dispense with the need to use an artificial diffusion-reduction factor at scale borders.
The results show that this scheme produces a scale-by-scale colouring and a CA behaviour, not only with 3D lattices of idealised hexagonal prisms, but also with realistic quasi-hexagonal lattices of 3D scales reconstructed from episcopic microscopy data. In other words, solution of the system of RD equations on the reconstructed actual 3D geometry of the skin scales produces a spatially-discrete pattern exactly superposed on the lattice of scales, validating the central assumption used in the previous study (Nature 2017). The Milinkovitch's team also shows now that the ‘snapping’ of the pattern to the borders of the scales is robust to variation of the RD-model, i.e., it occurs with vastly different sets of partial differential equations (PDEs).
They also propose a dimensionality-reduction approach that allows performing the simulations in 2D while integrating skin thickness as position-dependent RD components’ concentrations and effective diffusion coefficients. This scheme is slightly less accurate than bona fide 3D simulations but is more realistic than the reduction of diffusion coefficients along 1D edges of a 2D lattice of polygonal scales. Yet, they also show that the emergence of a CA behaviour is robust to alterations of the real skin geometry as all approaches generate very similar qualitative outcomes, i.e., non-trivial reduction of skin thickness at scale boundaries is sufficient for a transition between continuous and discrete patterns.
Finally, given that RD systems are known not to scale (i.e., the absolute length scale of the pattern should remain invariant), the team explored the effect of growth on the skin-colour patterning process in ocellated lizards. They show that the growth of the animal, across three years after hatching, deeply affects the dynamics of scale-colour change and produces a gradual decrease of the relative length scale of the pattern (measured in number of scale diameters) and a substantial increase of the average curvature of the pattern motifs. These results indicate that growth impacts form not only because of mechanical aspects of development but also in the emergence of anatomies controlled by RD-like systems.
Reference
Reaction-diffusion in a Growing 3D Domain of Skin Scales Generates a Discrete Cellular Automaton
Fofonjka & Milinkovitch
Nature Communications 2021
Contact: michel.milinkovitch@unige.ch
