Circadian locomotor behavior
Circadian rhythms
Circadian rhythms are the cyclic and persistent patterns of behavior and physiological processes exhibited by most organisms, ranging from cyanobacteria to human (Fig.1). These rhythms have a period of roughly 24 hours, matching the rotation of the Earth. Disruption of the circadian rhythms in humans causes sleep disorders and is also associated with many other health problems such as bipolar disorders and depression (1, 2).
Figure 1
At the molecular level, the core molecular clocks that present within many cells and tissues generate circadian rhythms. Accumulating evidence indicates that negative feedback loops of transcription are the design principle of the molecular clock core components in eukaryotes. Many clock genes are also conserved across a range of phylogenetic groups. These core clocks control rhythmic behavior and physiology principally by regulating rhythmic changes in downstream gene expression or protein activities (3, 4).
At the organismal level, the intrinsic periodicity is revealed as free-running rhythms when organisms are kept under constant conditions, such as constant darkness (DD). However, natural environments usually undergo daily changes such as light-dark cycles (LD) and organisms synchronize their circadian rhythms with these cycles, which is a process called “entrainment”. On the other hand, fluctuations in temperature have little impact on circadian rhythms. These seemingly opposing characteristics allow organisms to anticipate and prepare for regular and predictable environmental changes of the day, night and season. Components of the molecular clocks and the dedicated neural circuits are crucial to control both aspects of the circadian behavior.
The Drosophila circadian rhythms
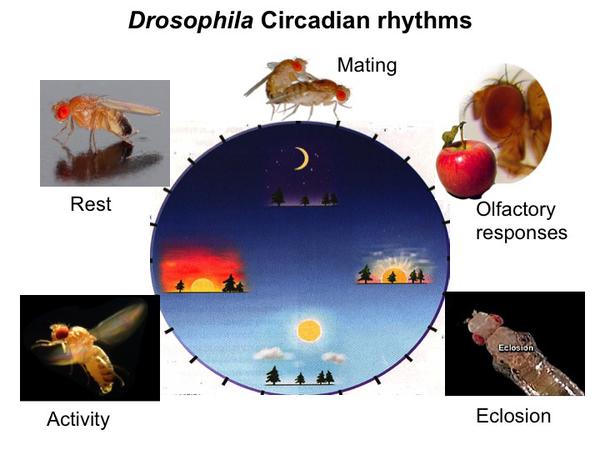
Figure 2
Drosophila displays circadian rhythms in various physiological and behavioral processes (Fig.2). Locomotor activity rhythms of flies show bimodal patterns, which peak at dawn and dusk in LD cycles. These rhythms sustain endlessly in DD. Approximately 150 clock-containing neurons in the adult brain make up the circuit controlling this circadian behavior. The clock neurons are classified into 7 subgroups based on their anatomical locations and characteristics. Developing animals also have fully functional, yet simpler clock circuits, which consist of only 3 groups of clock neurons (5). It has been suggested that neurons in the lateral brain (Lateral Neurons, LNs) contain the oscillators that control morning and evening activity (M-and E- cells) and therefore serve as the central pacemakers to generate rhythms. In contrast, other clock neurons appear to mediate input of the environmental information to the central pacemakers (6, 7). Recent studies have mapped the M-cells to the small ventral Lateral Neurons (s-LNvs) and E- cells to the dorsal Lateral Neurons (LNds) and some of Dorsal Neurons 1 (DN1) (8, 9). M-cells are not only required for the morning activity in LD, but also indispensable for driving rhythms in DD. Thus, many groups now interpret the term “M-cells” as “main oscillator” (10).
Research Projects
Despite the advances in circadian rhythms research, our understanding of the circadian circuit is still limited. In particular, roles of many clock neurons, organization of the circadian circuits and neurochemical basis of the neuronal communication among clock neurons remain poorly understood.
1. Molecular mechanisms of circadian circuit organization and operation
To understand the molecular underpinning of the circadian circuit organization and functioning, we have set out the molecular characterization of clock neuron subtypes. Using a novel technique to isolate and analyze RNA expression from a small number of specific neurons, we profiled genome-wide gene expression in several key subtypes of clock neurons (11, 12). We further showed that two nuclear receptor genes, unfulfilled (unf; DHR-51) and E75 play key roles in the functioning of the M-cells (13, 14). Our continuing efforts to dissect molecular mechanisms underlying the operation of circadian circuitry include identification of UNF and E75 targets and more comprehensive transcriptome analysis of clock neuron sub clusters.
2. Circadian neural circuit dynamics
Molecular rhythms are thought to control rhythmic neuronal activity and/or transmitter release. Conversely, there is a growing body of evidence that inter-neuronal signaling contributes to the synchronization and amplitude of the rhythms of clock neurons in both mammals and Drosophila (15, 16). Our goal is to decipher how inter-neuronal communication affects intracellular molecular clockwork, and how circuit-wide molecular and neuronal rhythms are integrated to generate rhythmic behavior. To this end, we developed fluorescence circadian reporter fly lines and live-imaging system using cultured brain and dissociated neurons. Our system allows for a spatiotemporally controlled manipulation of gene expression and neuronal activity together with the real-time recording of molecular oscillation in clock neurons.
Parkinson’s disease
Parkinson’s disease (PD) is the movement disorder characterized by the locomotor defects such as tremor, bradykinesia, rigidity and postural instability, affecting over 1% of the global population over 60 years of age. Motor symptoms of PD primarily arise from the progressive loss of dopaminergic (DA) neurons in the substantia nigra (SN). Despite the advances in gene discovery associated with familial PD, the knowledge of the PD pathogenesis is still limited. In particular, why the degeneration is specific to DA neurons and why it is progressive remain enigmatic. Lack of animal models that show genuinely progressive DA neuron degeneration has also hindered the study on this central issue.
Overall goal of our research in this topic is to understand the mechanisms underlying selective and progressive degeneration of the DA neurons. We will address these central questions using a novel Drosophila PD model we have established, and later by generating new mouse models.
Research Projects
We have recently established a novel PD model in Drosophila that offers an unusual and exciting opportunity to address the mechanisms underlying selective and progressive degeneration of DA neurons (17). Our model flies - the Fer2 (48-related-2) gene loss-of-function mutants - show specific and progressive death of brain DA neurons, severe locomotor defects and reduced life span (Movie 1, 2). We further showed that degeneration of DA neurons in Fer2 loss-of function mutants coincides with the systemic increase in reactive oxygen species (ROS) and mitochondrial dysfunction within DA neurons. Because increased ROS production and mitochondrial dysfunctions are pathological hallmarks of PD, our results underscore that Fer2 mutants recapitulate cellular and organismal characteristics of PD (17).
Encouraged by these exciting results, we are investigating upstream and downstream pathways of Fer2 to understand molecular mechanisms contributing to the survival of DA neurons. Furthermore, based on the knowledge gained from our work using Fer2 mutant flies, we are generating novel mouse models of PD.
Video 1: Brain DA neurons in the control flies
Video 2: Brain DA neurons in the Fer21 mutant flies
References
- Benca R, et al. (2009) "Biological rhythms, higher brain function, and behavior: gaps, opportunities and challenges". Brain Res Rev.
- Stevens RG, et al. (2007) Meeting report: the role of environmental lighting and circadian disruption in cancer and other diseases. Environ Health Perspect 115(9):1357-1362.
- Hastings MH, Maywood ES, & O'Neill JS (2008) Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18(17):R805-R815.
- Yu W & Hardin PE (2006) Circadian oscillators of Drosophila and mammals. J Cell Sci 119(Pt 23):4793-4795.
- Helfrich-Forster C (2005) Neurobiology of the fruit fly's circadian clock. Genes Brain Behav 4(2):65-76.
- Grima B, Chelot E, Xia R, & Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431(7010):869-873.
- Stoleru D, Peng Y, Agosto J, & Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431(7010):862-868.
- Helfrich-Forster C, et al. (2007) The lateral and dorsal neurons of Drosophila melanogaster: new insights about their morphology and function. Cold Spring Harb Symp Quant Biol 72:517-525.
- Picot M, Cusumano P, Klarsfeld A, Ueda R, & Rouyer F (2007) Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol 5(11):e315.
- Nitabach MN & Taghert PH (2008) Organization of the Drosophila circadian control circuit. Curr Biol 18(2):R84-93.
- Kula-Eversole E, et al. (2010) Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A 107(30):13497-13502.
- Nagoshi E, et al. (2010) Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 13(1):60-68.
- Beuchle D, Jaumouille E, & Nagoshi E (2012) The Nuclear Receptor unfulfilled Is Required for Free-Running Clocks in Drosophila Pacemaker Neurons. Curr Biol 22(13):1221-1227.
- Jaumouille E, Machado Almeida P, Stahli P, Koch R, & Nagoshi E (2015) Transcriptional regulation via nuclear receptor crosstalk required for the Drosophila circadian clock. Curr Biol 25(11):1502-1508.
- Freeman GM, Jr., Krock RM, Aton SJ, Thaben P, & Herzog ED (2013) GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron 78(5):799-806.
- Mizrak D, et al. (2012) Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Curr Biol 22(20):1871-1880.
- Bou Dib P, et al. (2014) A Conserved Role for p48 Homologs in Protecting Dopaminergic Neurons from Oxidative Stress. Plos Genet 10(10).
