- news
- 27-08-2010
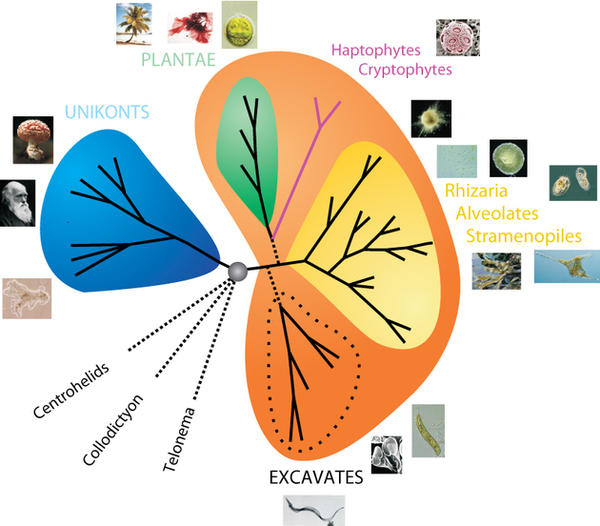
The use of genomic data in a phylogenetic context allows one to infer evolutionary relationships between groups of organisms that were impossible to tackle a few years back on a single-gene basis. Recently several Expressed Sequence Tag (EST) datasets and full genomes have been made freely available, allowing the construction of huge alignments containing more than a hundred genes for species representing almost all large groups of eukaryotes. It is an exciting time for scientists working in phylogenomics since it is becoming possible to generate great amounts of data for key species, thank to new sequencing strategies (eg. 454 Sequencing). Thus, the remaining uncertainties in the tree of eukaryotes are being rapidly addressed by several laboratories.
One of these uncertainties has been addressed by Fabien Burki, member of Pr Jan Pawlowski's laboratory, with help of a colleague from the University of Oslo, in a forthcoming paper recently accepted in Biology Letters. In this paper, Fabien and colleagues investigated ancient evolution among the major eukaryotic supergroups, by using the broadest multigene dataset to date (65 species, 135 genes). Their results provide strong support for the clustering of nearly all photosynthetic lineages and raise the question of a possible unique origin of plastids (organnelles responsible for photosynthesis in plants). They speculate that the high observable diversity of plastids within a single assemblage can be traced back to its last common ancestor. At its deepest level, the tree of eukaryotes now receives strong support for only two monophyletic megagroup comprising most of the eukaryotic diversity.
